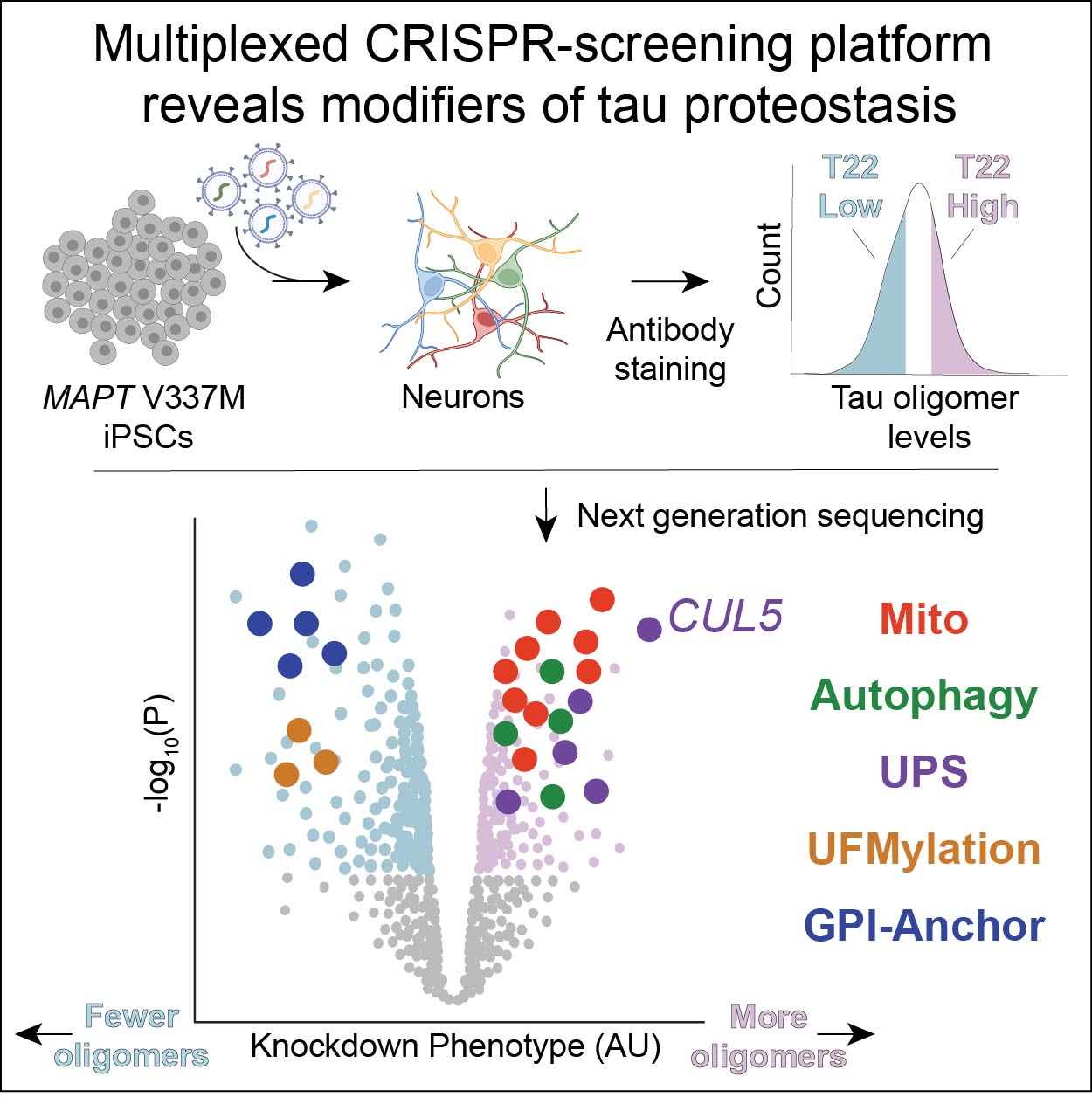
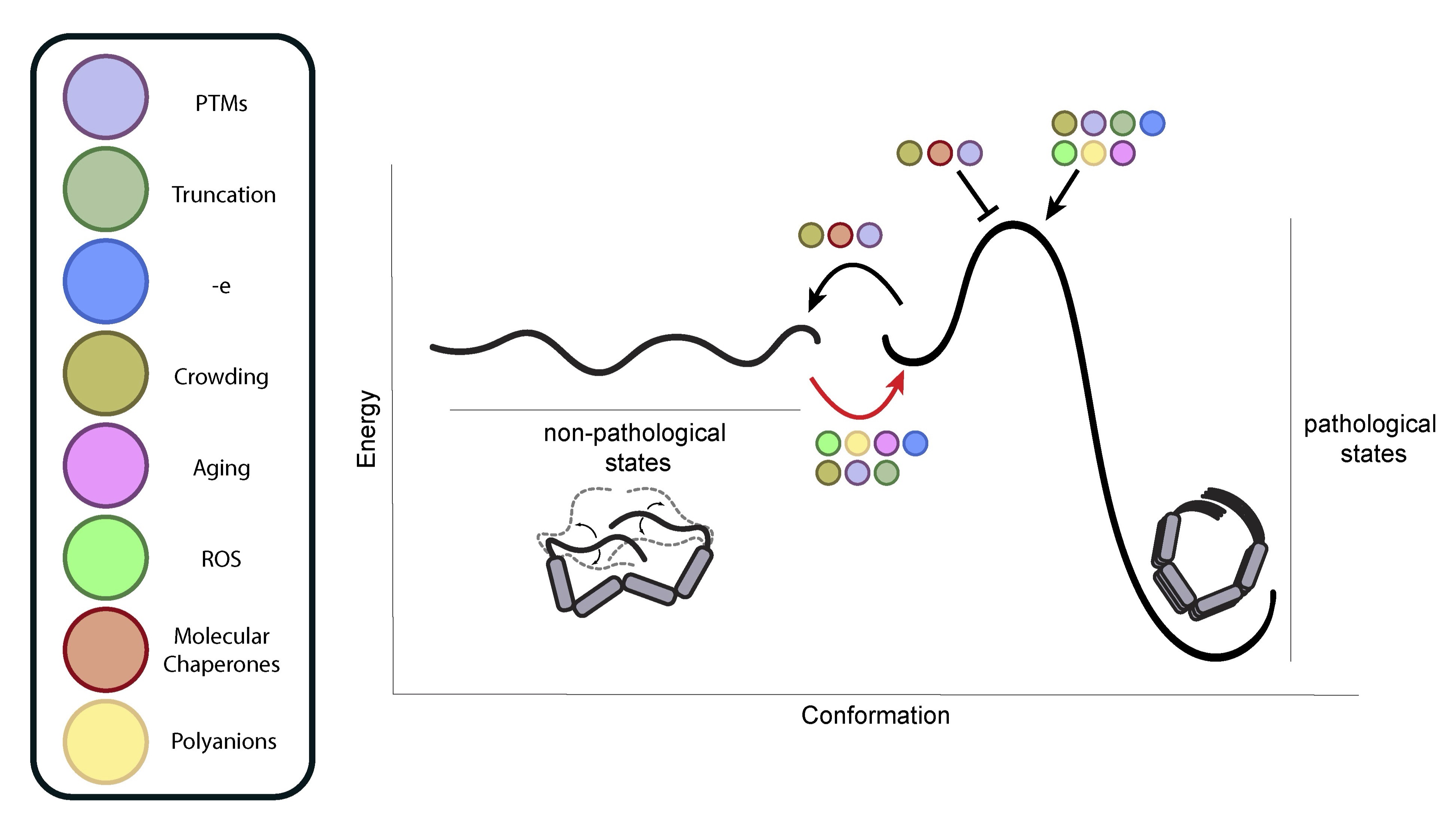
CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis.
Samelson AJ, Ariqat N, McKetney J, Rohanitazangi G, Bravo CP, Bose RS, Travaglini KJ, Lam VL, Goodness D, Dixon G, Marzette E, Jin J, Tian R, Tse E, Abskharon R, Pan H, Carroll EC, Lawrence RE, Gestwicki JE, Rexach RE, Eisenberg DS, Kanaan NM, Southworth DR, Gross JD, Gan L, Swaney DL, Kampmann M
Cell,
2026
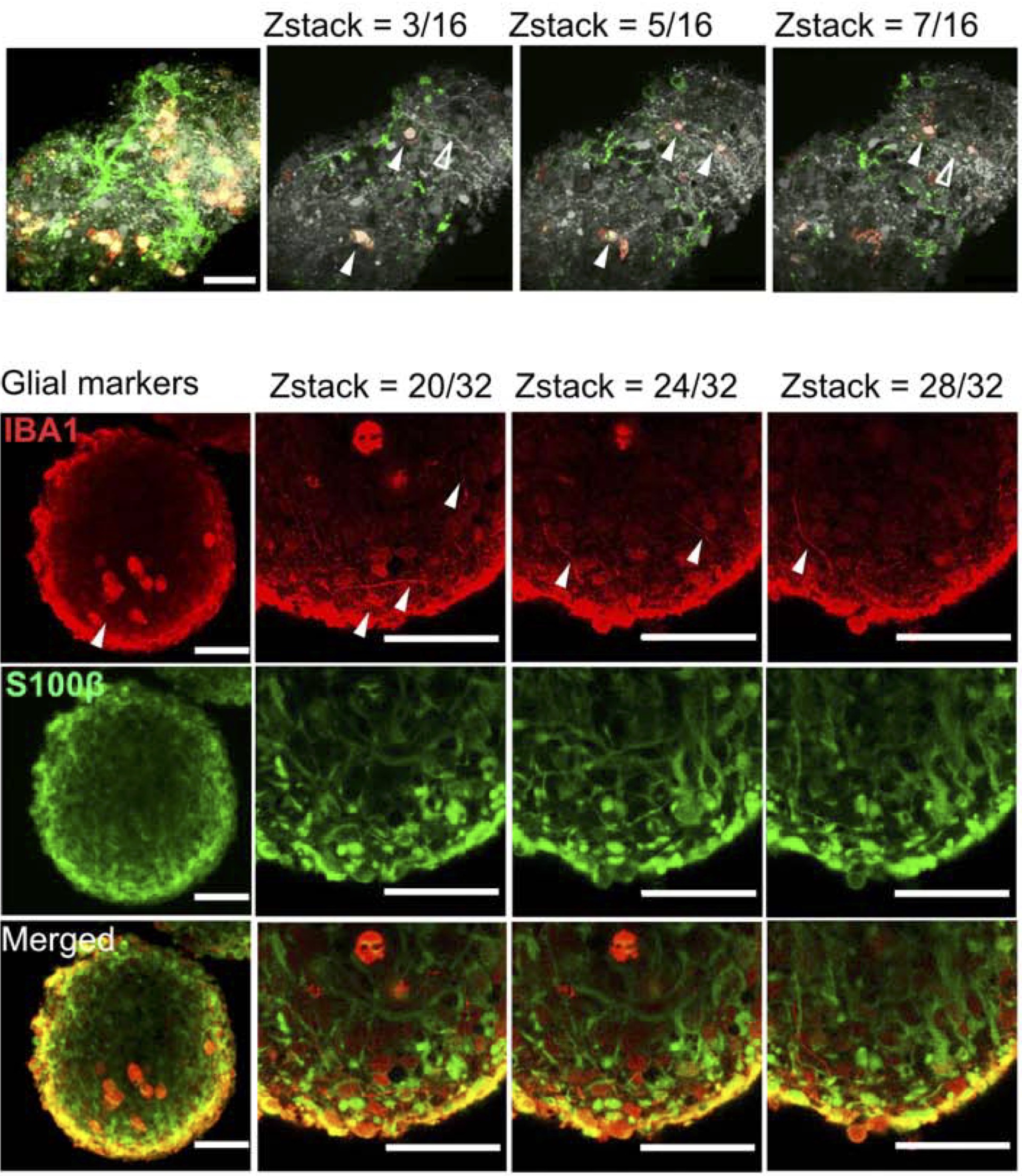
CRISPRi-based screens in iAssembloids to elucidate neuron-glia interactions.
Li E, Benitez C, Boggess SC, Koontz M, Rose IVL, Martinez D, Draeger N, Teter OM, Samelson AJ, Pierce N, Ullian EM, Kampmann M.
Neuron,
2025
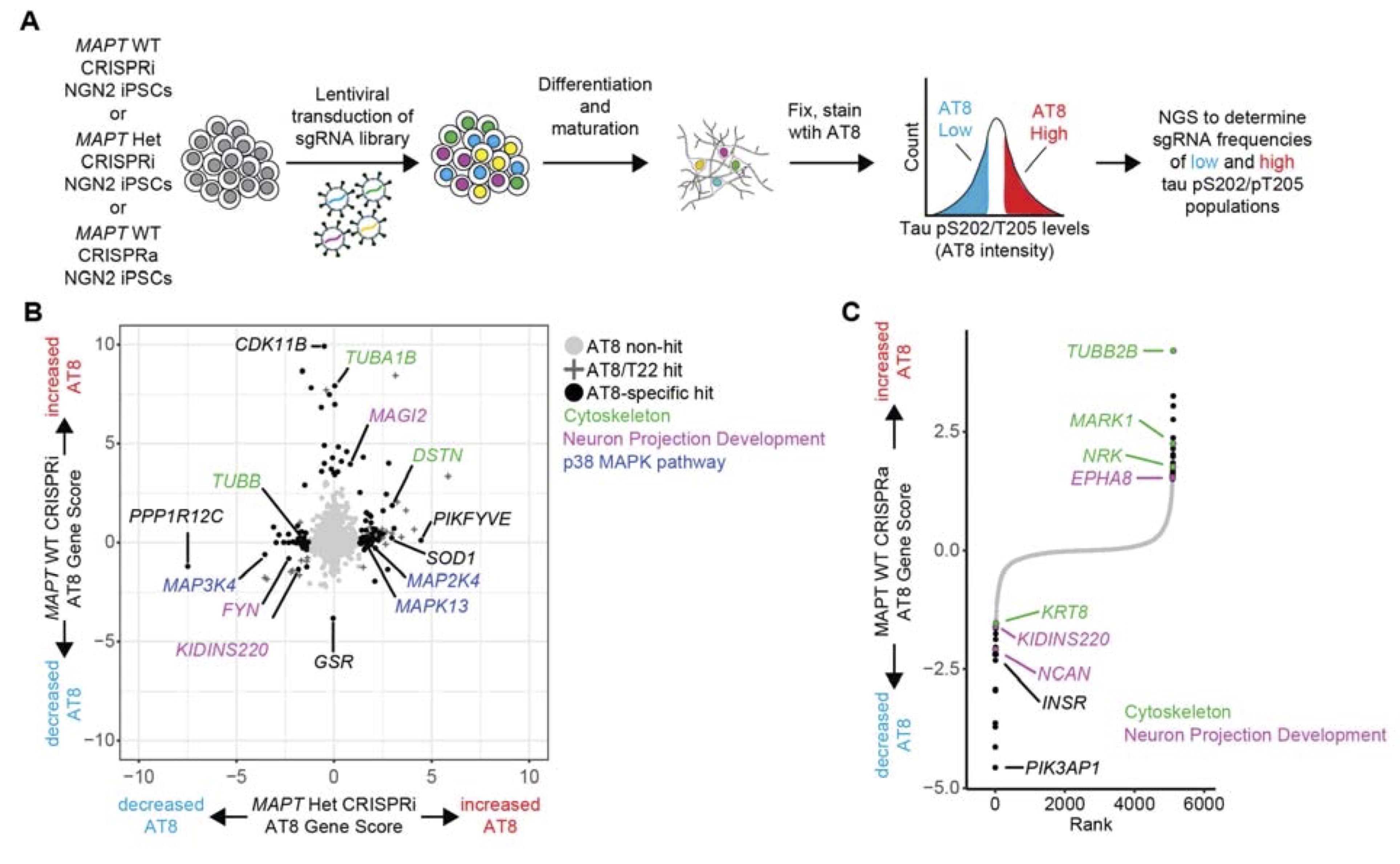
The disease-causing tau V337M mutation induces tau hypophosphorylation and perturbs axon morphology pathways.
Mohl GA, Dixon G, Marzette E, McKetney J, Samelson AJ, Serras CP, Jin J, Li A, Boggess SC, Swaney DL, Kampmann M.
Biorxiv,
2024
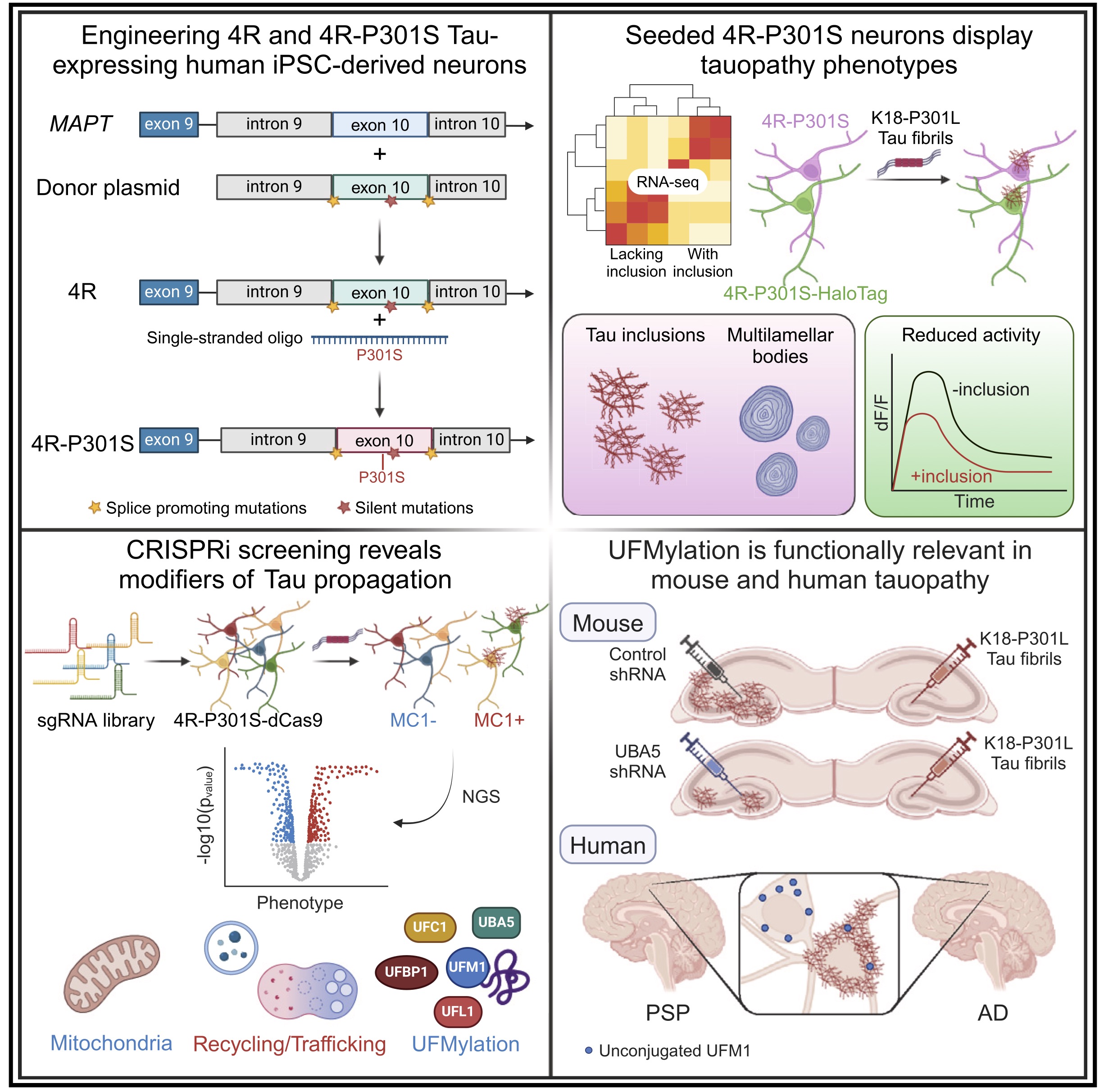
Human iPSC 4R tauopathy model uncovers modifiers of tau propagation.
Parra Bravo C, Giani AM, Madero-Perez J, Zhao Z, Wan Y, Samelson AJ, Wong MY, Evangelisti A, Cordes E, Fan L, Ye P, Zhu D, Pozner T, Mercedes M, Patel T, Yarahmady A, Carling GK, Sterky FH, Lee VMY, Lee EB, DeTure M, Dickson DW, Sharma M, Mok SA, Luo W, Zhao M, Kampmann M, Gong S, Gan L.
Cell,
2024
The Cellular Environment Guides Self-Assembly and Structural Conformations of Microtubule-Associated Protein Tau (MAPT)
Montgomery KM, Samelson AJ, Gestwicki JE
Il. J. Chem.,
2023
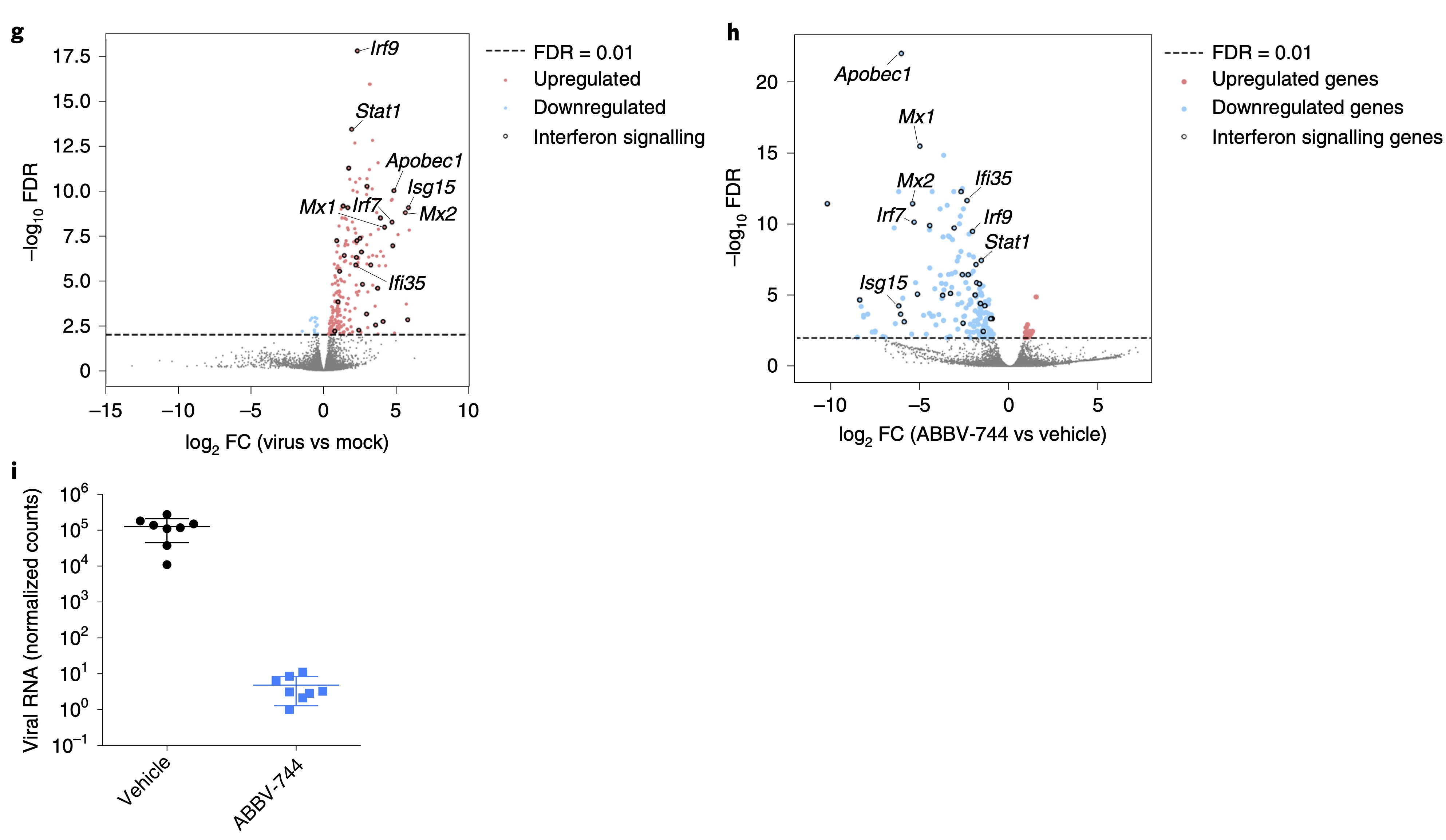
BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2.
Samelson AJ, Tran QD, Robinot R, Carrau L, Rezelj VV, Kain AM, Chen M, Ramadoss GN, Guo X, Lim SA, Lui I, Nuñez JK, Rockwood SJ, Wang J, Liu N, Carlson-Stevermer J, Oki J, Maures T, Holden K, Weissman JS, Wells JA, Conklin BR, TenOever BR, Chakrabarti LA, Vignuzzi M, Tian R, Kampmann M
Nature Cell Biology,
2022
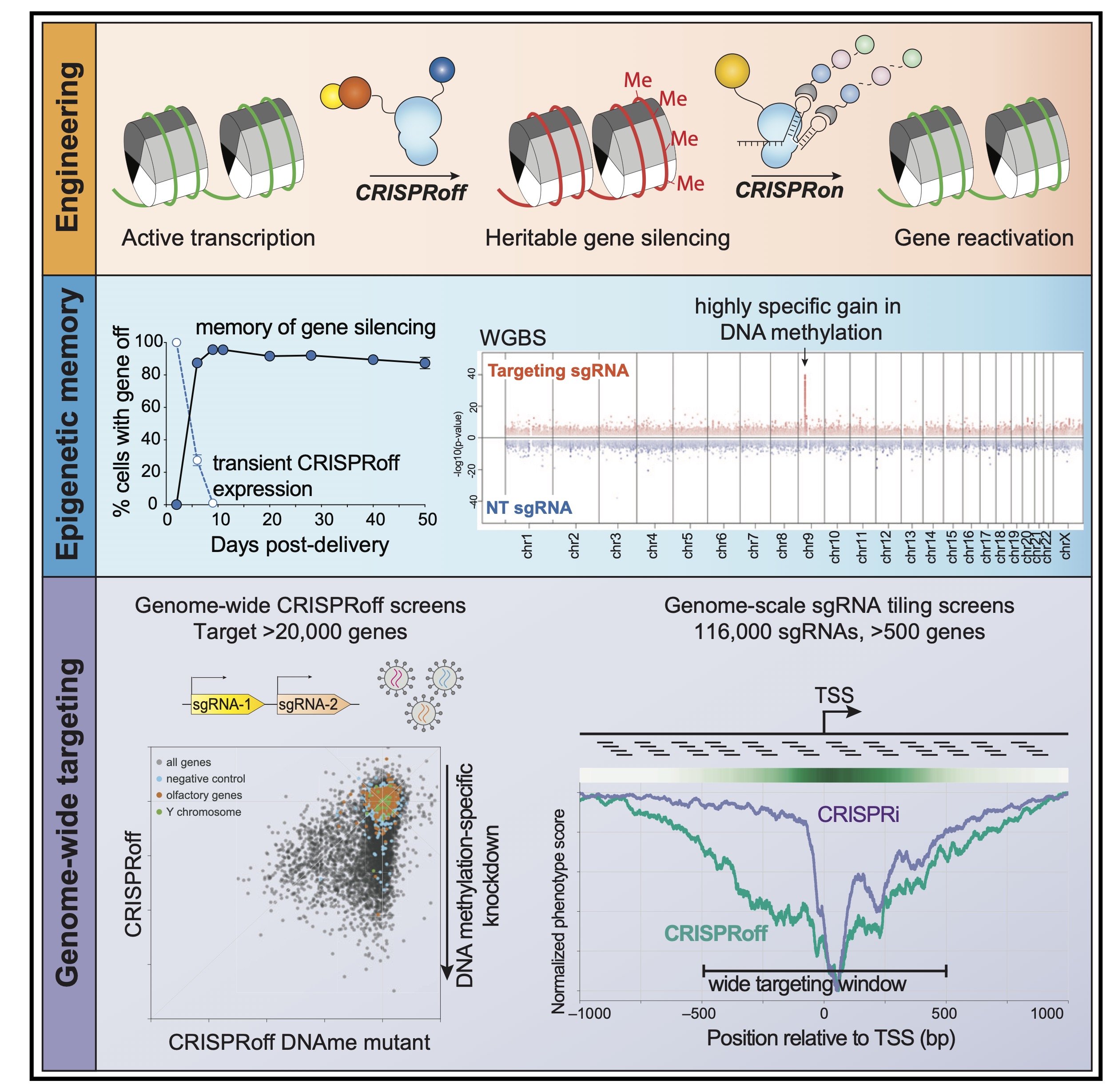
Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing.
Nuñez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, Ramadoss GN, Shi Q, Hung KL, Samelson AJ, Pogson AN, Kim JYS, Chung A, Leonetti MD, Chang HY, Kampmann M, Bernstein BE, Hovestadt V, Gilbert LA, Weissman JS.
Cell,
2021
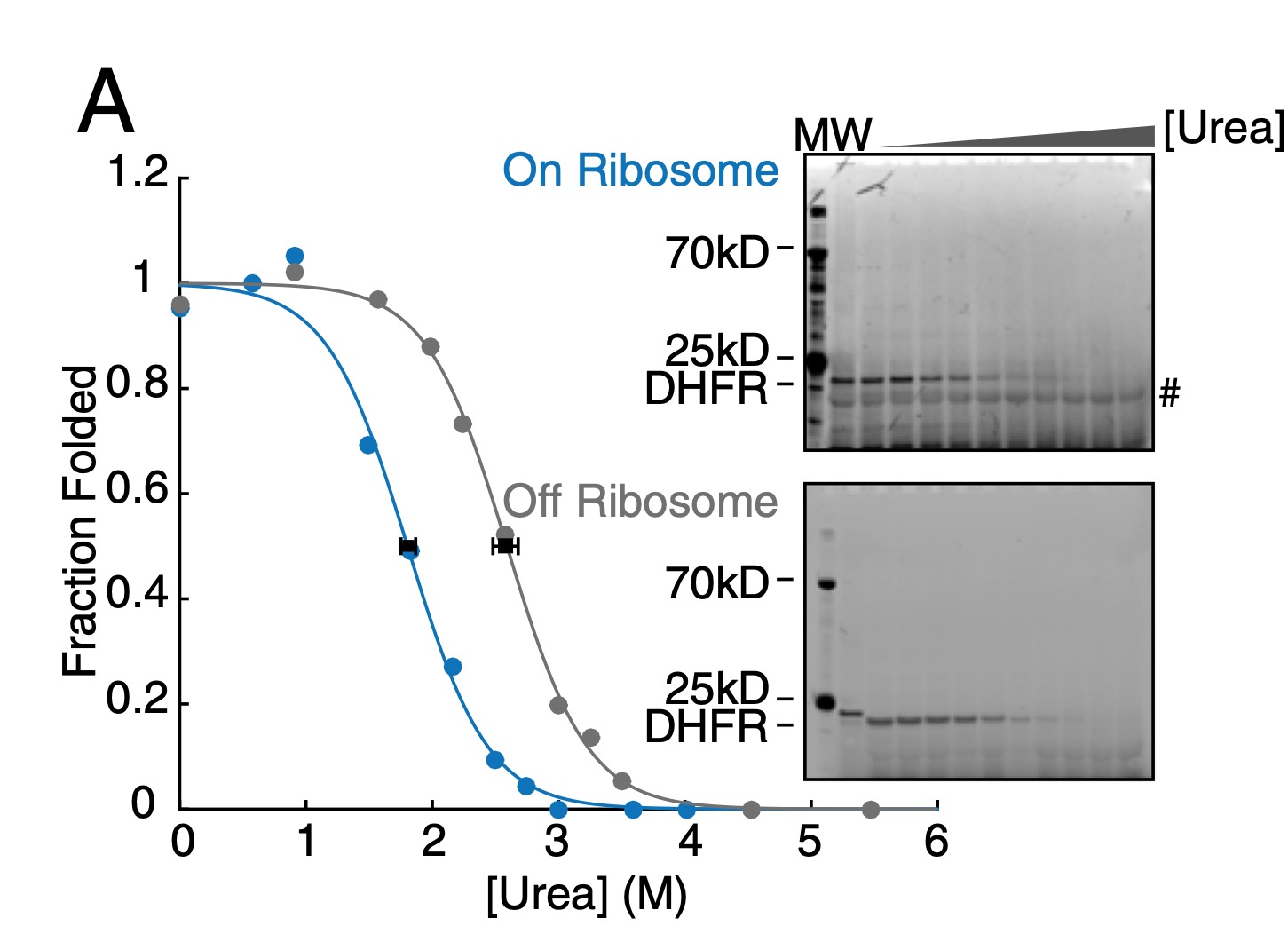
The folding and unfolding behavior of ribonuclease H on the ribosome
Jensen MK, Samelson AJ, Steward A, Clarke J, Marqusee S
JBC,
2020
Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Wolff AM, Young ID, Sierra RG, Brewster AS, Martynowycz MW, Nango E, Sugahara M, Nakane T, Ito K, Aquila A, Bhowmick A, Biel JT, Carbajo S, Cohen AE, Cortez S, Gonzalez A, Hino T, Im D, Koralek JD, Kubo M, Lazarou TS, Nomura T, Owada S, Samelson AJ, Tanaka R, Tanaka T, Thompson EM, van den Bedem H, Woldeyes RA, Yumoto F, Zhao W, Tono K, Boutet S, Iwata S, Gonen T, Sauter NK, Fraser JS, Thompson MC
IUCrJ,
2020
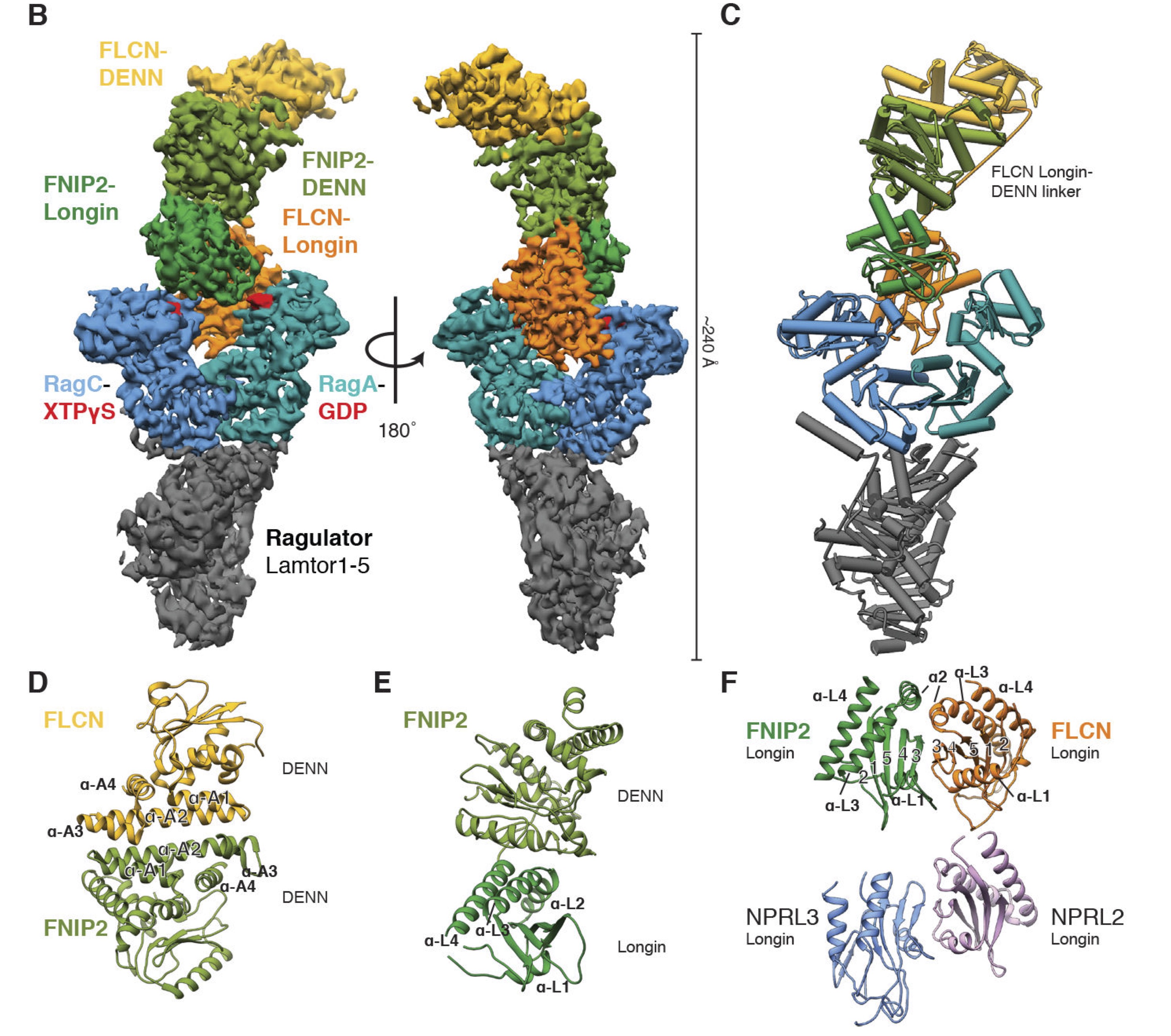
Structural Mechanism of a Rag GTPase Activation Checkpoint by the Lysosomal Folliculin Complex
Lawrence RE, Fromm SA, Fu Y, Yokom AL, Kim DJ, Thelen AM, Young LN, Lim C-Y, Samelson AJ Hurley JH, Zoncu R
Science,
2019
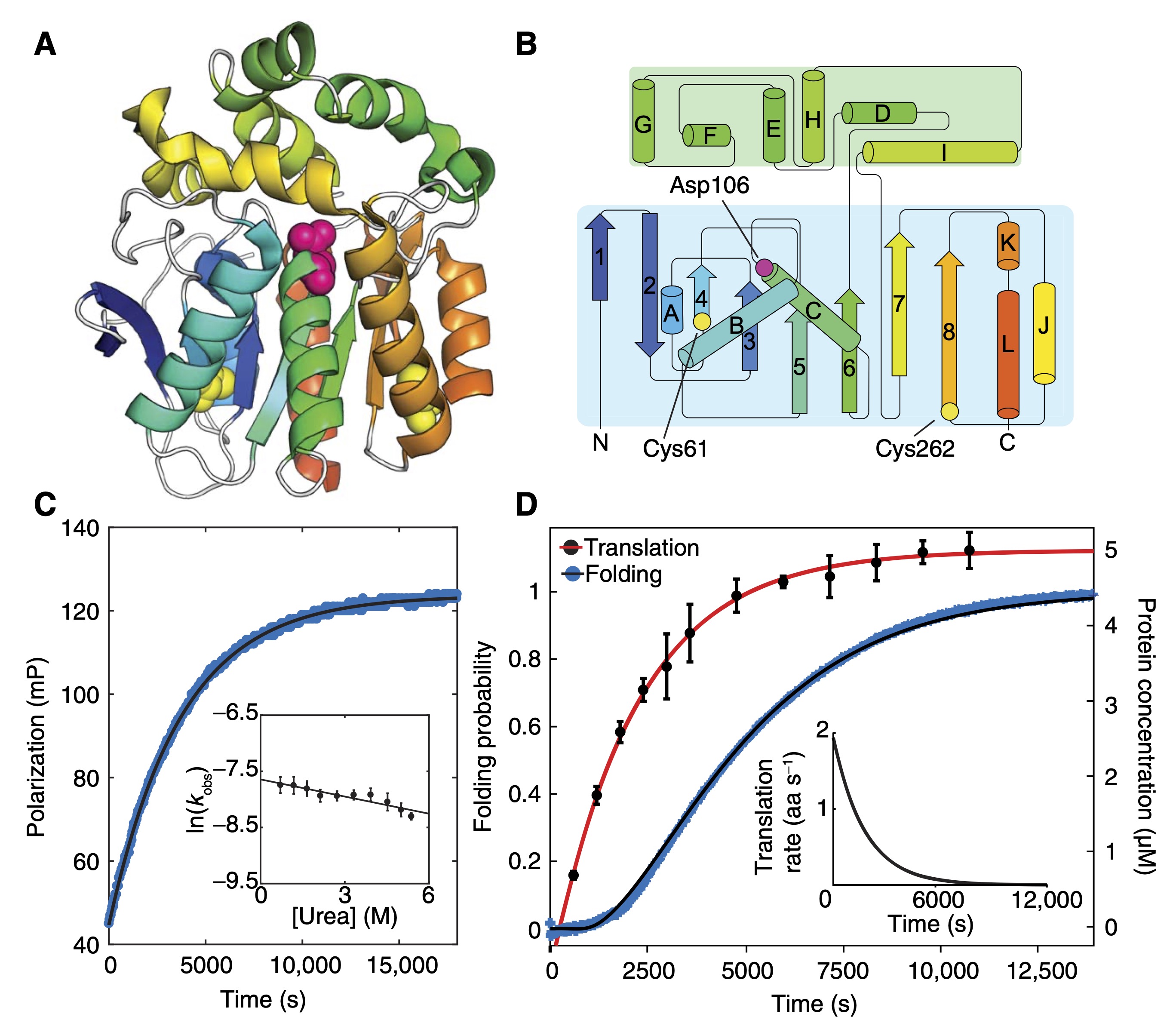
Kinetic and structural comparison of a protein’s cotranslational folding and refolding pathways
Samelson AJ, Bolin E, Costello SM, Sharma AK, O’Brien EP, Marqusee S
Science Advances,
2018
Quantitative determination of ribosome nascent chain stability
Samelson AJ, Jensen MK, Soto RA, Cate JH, Marqusee S
PNAS,
2016
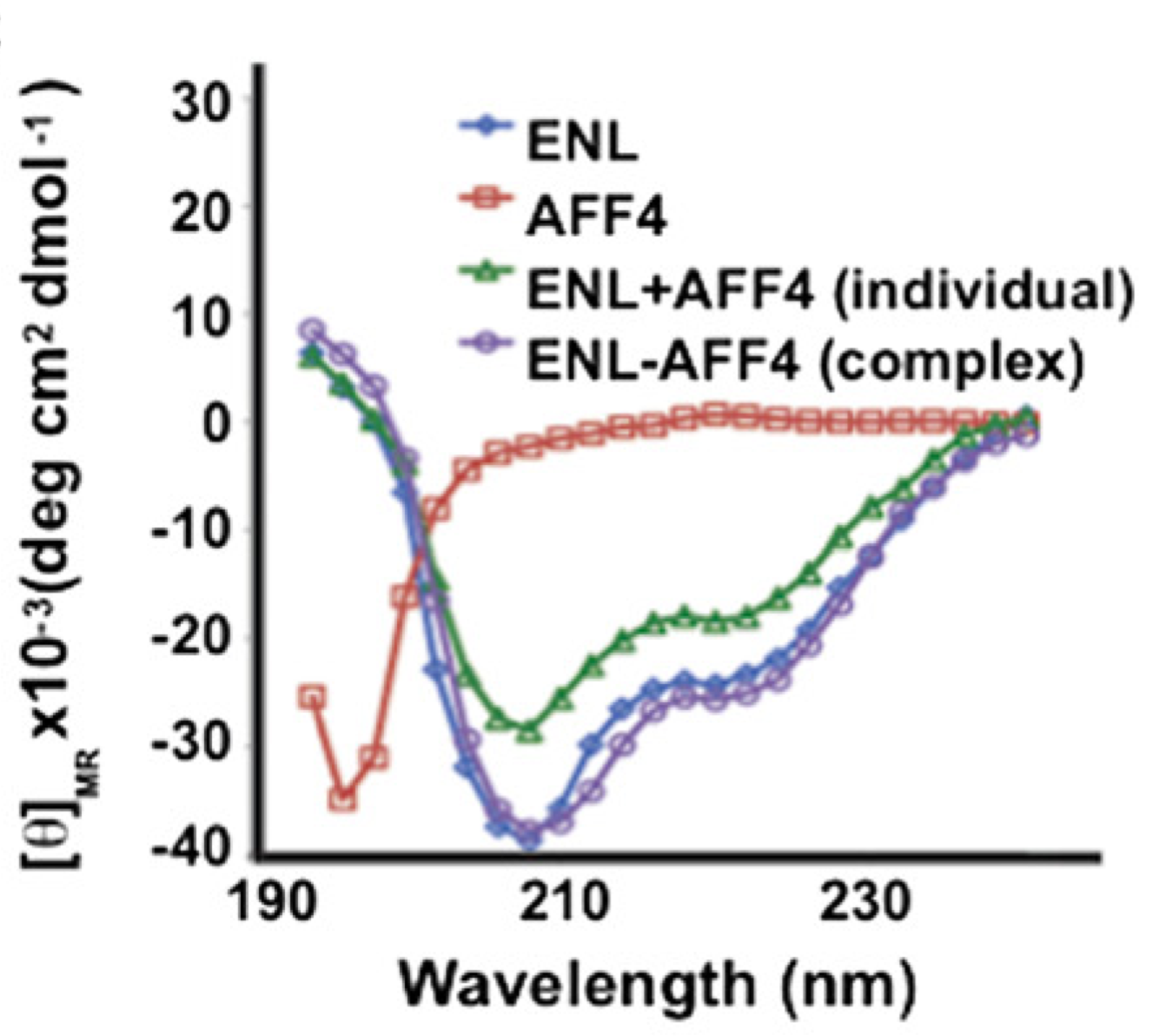
HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold
Chou S, Upton H, Bao K, Schulze-Gahmen U, Samelson AJ, He N, Nowak A, Lu H, Krogan NJ, Zhou Q, Alber T
PNAS,
2013
Accessing protein conformational ensembles by room-temperature X-ray crystallography
Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T
PNAS,
2011